Abstract
Background
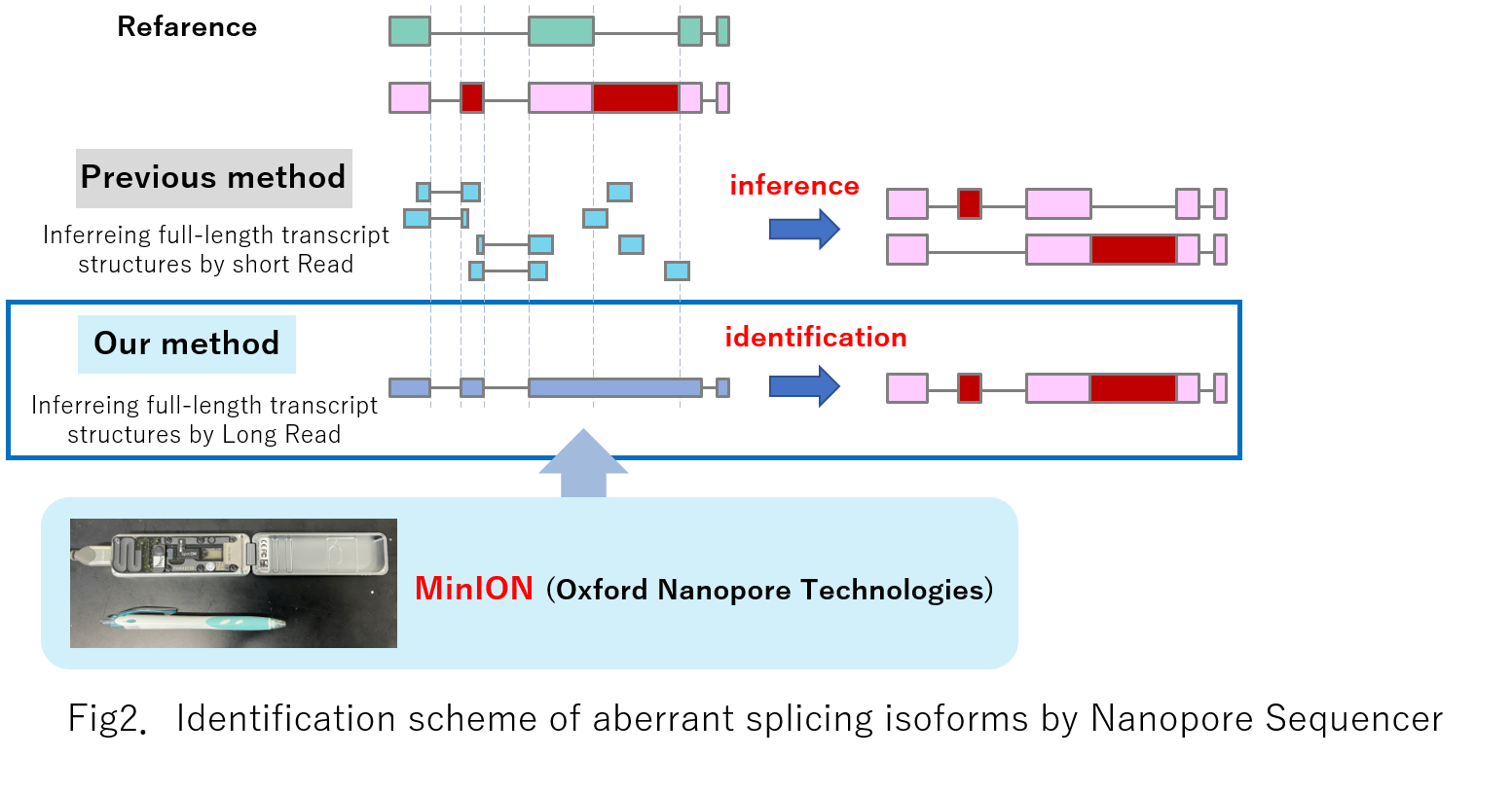
Long-read sequencing of full-length cDNAs enables the detection of structures of aberrant splicing isoforms in cancer cells. These isoforms are occasionally translated, presented by HLA molecules, and recognized as neoantigens. This study used a long-read sequencer (MinION) to construct a comprehensive catalog of aberrant splicing isoforms in non-small-cell lung cancers, by which novel isoforms and potential neoantigens are identified.Results
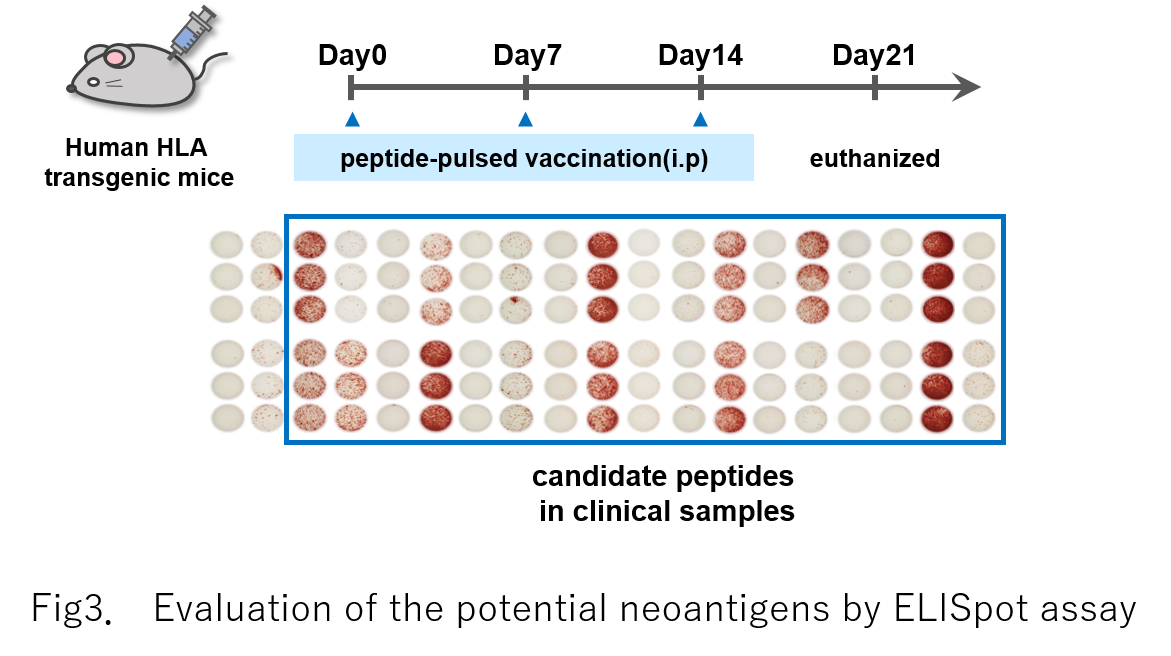
Full-length cDNA sequencing is performed using 22 cell lines, and a total of 2021 novel splicing isoforms are identified. The protein expression of some of these isoforms is then validated by proteome analysis. Ablations of a nonsense-mediated mRNA decay (NMD) factor, UPF1, and a splicing factor, SF3B1, are found to increase the proportion of aberrant transcripts. NetMHC evaluation of the binding affinities to each type of HLA molecule reveals that some of the isoforms potentially generate neoantigen candidates. We also identify aberrant splicing isoforms in seven non-small-cell lung cancer specimens. An enzyme-linked immune absorbent spot assay indicates that approximately half the peptide candidates have the potential to activate T cell responses through their interaction with HLA molecules. Finally, we estimate the number of isoforms in The Cancer Genome Atlas (TCGA) datasets by referring to the constructed catalog and found that disruption of NMD factors is significantly correlated with the number of splicing isoforms found in the TCGA-Lung Adenocarcinoma data collection.Conclusions
Our results indicate that long-read sequencing of full-length cDNAs is essential for the precise identification of aberrant transcript structures in cancer cells.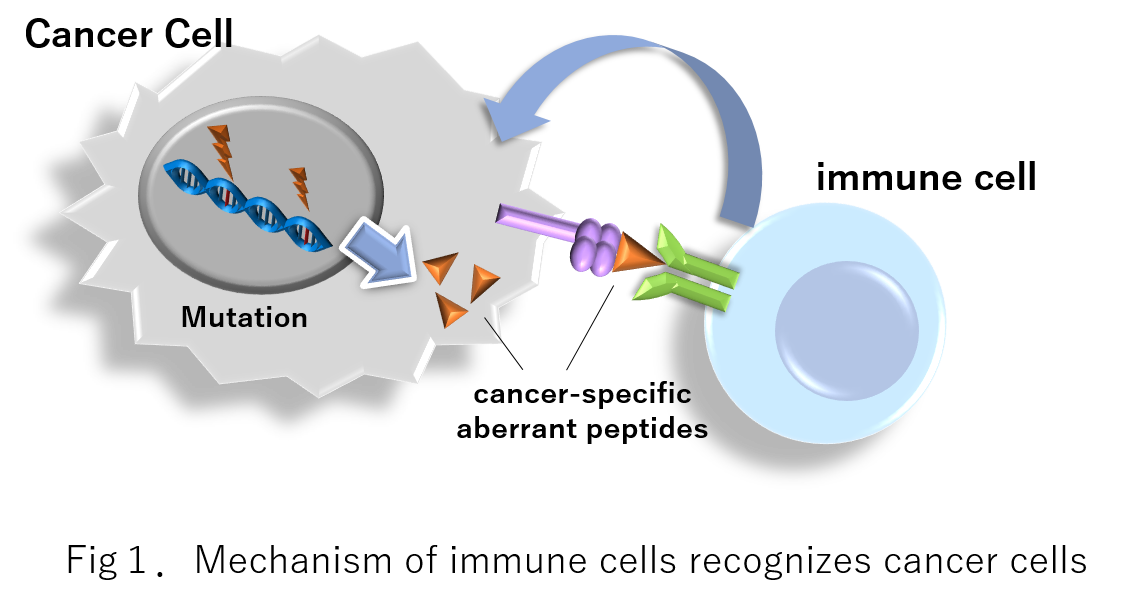
Accession:
Cell Line:
DRA010214,
DRA010215
Clinical Sample:
JGAS000245 * Controlled Access.
Cancer LongRNA Dataset
We prepared Cancer Specific Aberrant splicing isoforms list(22 Cell line, 7 Clinical samples )
・
Aberrant splicing isoforms list (.gtf file) of 22 Cell line
・
Aberrant splicing isoforms list (.gtf file) of 7 Clinical Samples
・Readme
The genome version is GRCh38.
File format (gtf)
Column 1: ChromosomeColumn 2: Source (MinION)
Column 3: Feature (transcript = whole transcript, exon = each exon)
Column 4: Start position
Column 5: End position (1-based)
Column 6: No Data
Column 7: Strand
Column 8: No Data
Column 9: Attribute (The detail is described below.)
Column 10: RefSeq accession number (RefSeq transcript that is the most similar to the isoform)
Column 11: Sample names in which the isoform was detected. (for merged file only)
Attribute format
1. gene_id : Not Entrez gene ID!! (The number for each gene used for the analysis internally.)2. trancript_id : The isoform ID used for the analysis internally. "gene symbol"_"chromosome"_"start positions for each exon junctions"+"end positions for each exon junctions (0-based)"_"isoform label"
3. gene_name : Gene symbol.
4. label : isoform_RefSeq: All junctions in the isoform are the same as those of the RefSeq transcript, but the combination is different.
isoform_nonRefSeq: At least one junction in the isoform is distinct from those of the RefSeq transcript.
5. exon_number : The number of exon in each isoform.